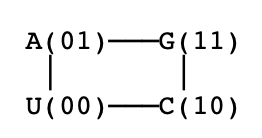The Result
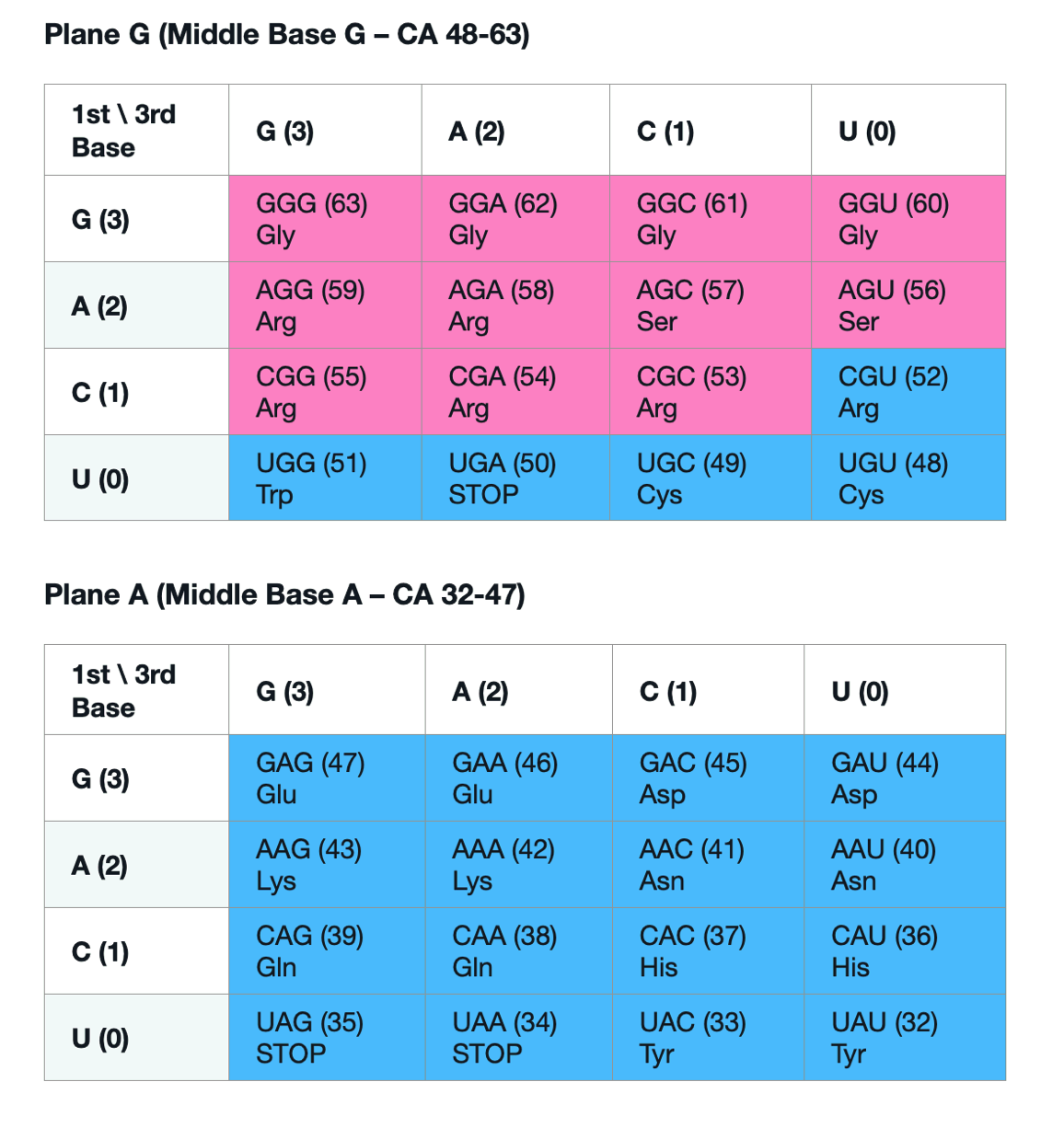
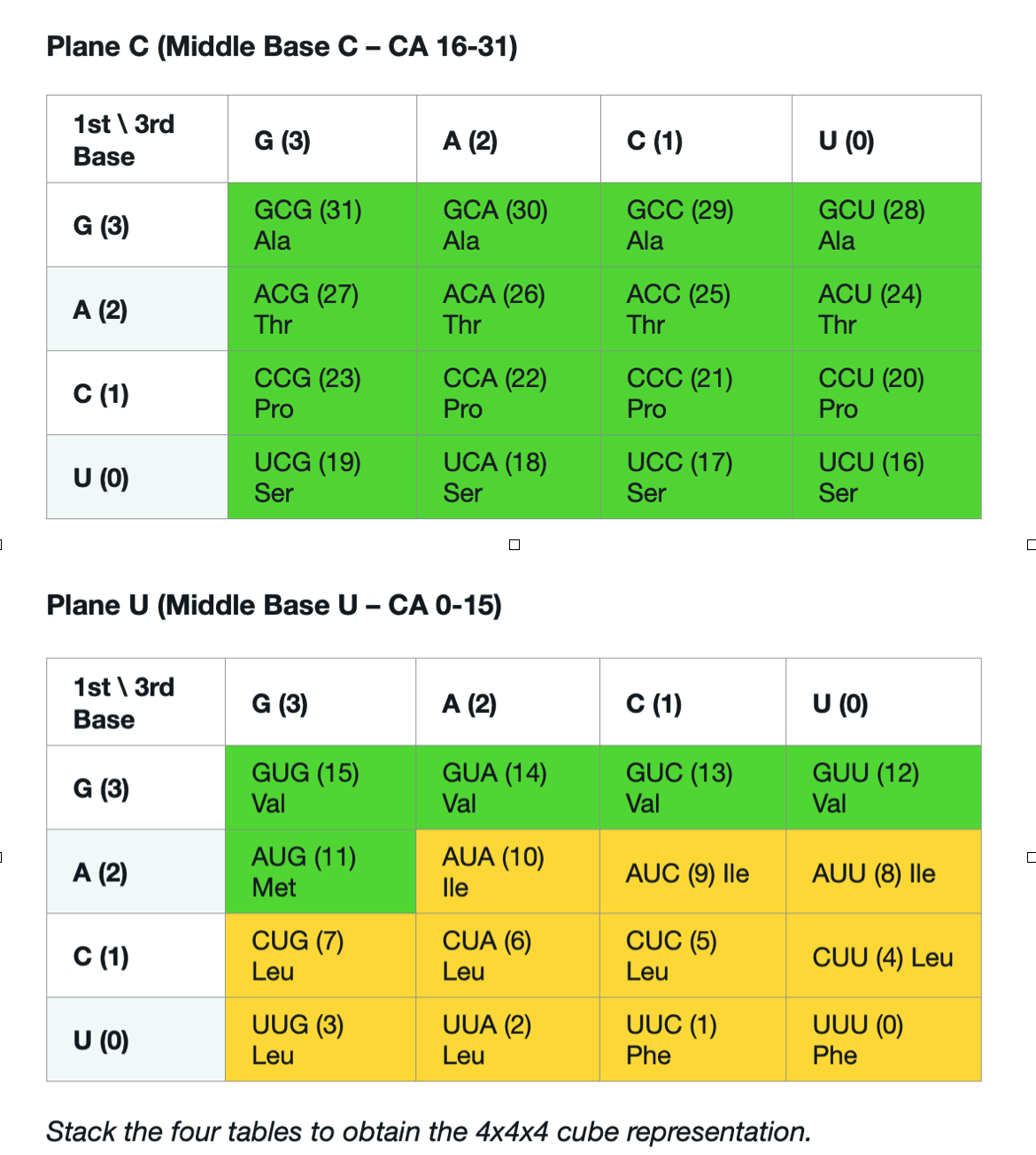
UCAG 4×16×1 generates a three-dimensional coordinate system organizing all 64 codons as a 4×4×4 cube
The Cube Structure
Every codon maps to coordinates 0-63. Four homo-nucleotide codons (UUU, CCC, AAA, GGG) anchor the main diagonal at positions 0, 21, 42, 63.
Single nucleotide mutations produce predictable coordinate changes: ΔCA = ±1, ±4, or ±16 single-digit changes in quaternary notation.
19 of 20 amino acids have all codons within a single plane (same middle nucleotide). Natural boundaries at 10/11, 31/32, and 52/53 create four functional domains.
Self-Referential Coordinates
Each codon's middle nucleotide sets its own reference. Flanking positions are measured against it without external frame required.
Domain Formation
Chemistry Domain (32-52): 50% of amino acids, 100% of STOP codons, all charged/aromatic residues. Probability by chance: <0.002%
Predictive Power
Coordinate distances correlate with biology: pathogenic mutations average ΔCA=17.3 vs benign at ΔCA=8.1
Species Validation
Bacterial codon usage shows systematic preferences for CGU(52) exactly where the framework predicts optimization, at the Chemistry/Adaptation boundary.


The Generating Template
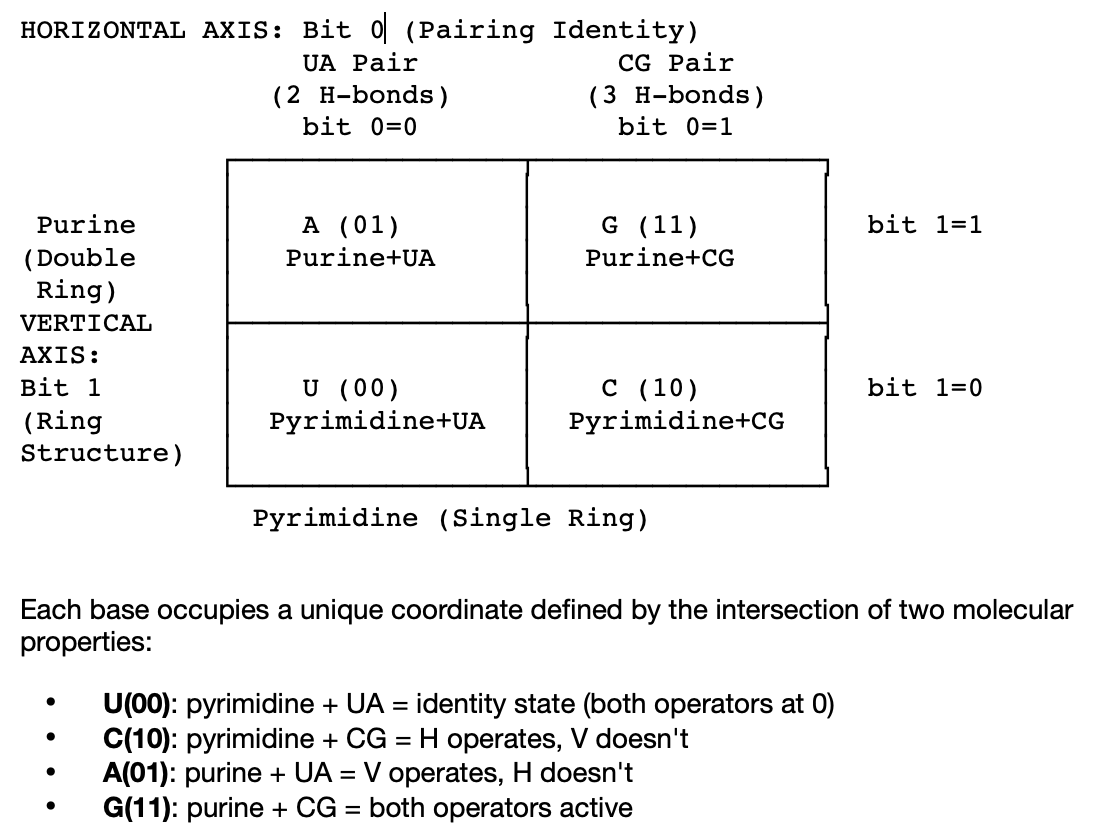
The entire 4×4×4 cube reduces to a single mathematical object: a 2×2 Gray code iterated three times. Two molecular properties, ring structure (pyrimidine/purine) and pairing identity (UA/CG) define two binary orthogonal axes that place each nucleotide at a unique coordinate.
Traversing the square changes exactly one bit per step: a Gray code. Within chemical families (U↔C, A↔G), single-bit adjacency provides error tolerance. Crossing the C|A boundary flips both bits: an irreversible two-bit regime transition. This template, applied at each of three codon positions with the weights 4×16×1 generates the complete 64-state space. No new mathematics emerges at scale.